tl;dr
- You can now do mediation and moderation analyses in jamovi and R with
medmod - Use
medmodfor an easy transition tolavaan
Introducing medmod
The PROCESS macro has been a very popular add-on for SPSS that allows you to do a wide variety of path model analyses, of which mediation and moderation analysis are probably the most well-known. We’ve received a lot of requests to add these analyses to jamovi, and today we are happy to announce the medmod module which allows you to do mediation and moderation analyses in jamovi (and R). In this blog post we will give a brief demonstration of the mediation analysis in medmod in both jamovi and R, and then show how you can use medmod in combination with lavaan, a powerful R package for latent variable modeling.
Mediation analysis
Mediation analysis allows you to explore whether a mediating variable can explain the relationship between two variables. For example, you might want to know whether the relationship between ‘personality similarity’ and ‘marital satisfaction’ is mediated by ‘shared activities’. Having similar personalities could lead to more time spend together doing shared activities, which in turn could lead to more marital satisfaction. If this is the case, decreasing the influence of ‘personality similarity’ on ‘shared activities’ (i.e., spending time doing shared activities regardless of having similar personalities or not) should decrease (or even nullify) the original relationship between ‘personality similarity’ and ‘marital satisfaction’.
The path model below illustrates the mediation model. Here, X is the predictor, M the mediator, and Y the outcome variable. While path c describes the direct effect of the predictor variable on the outcome variable, paths a and b together describe the indirect or mediated effect. If there is both an indirect and direct effect, it’s called partial mediation. If there is an indirect effect but no direct effect, it’s called full moderation.
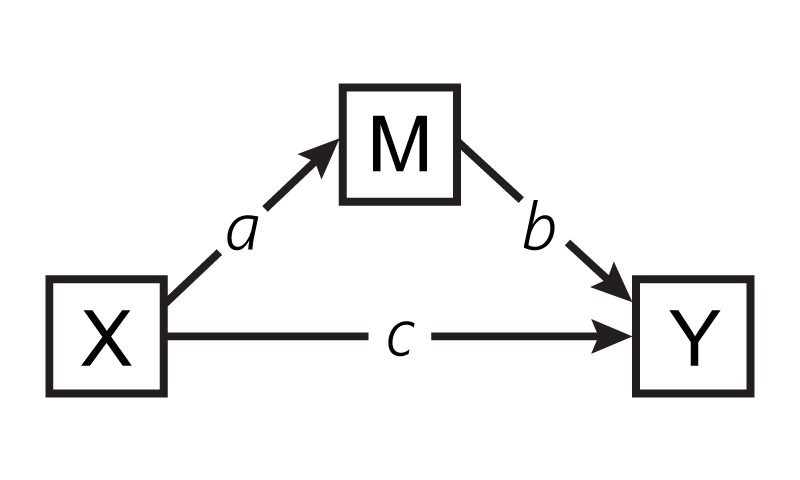
To demonstrate this analysis, we will use the data set from Pollack, VanEpps, and Hayes (2012), who explored whether the relationship between entrepreneurs withdrawing from entrepreneurial activities and financial stress is mediated by them experiencing feelings of depression. Did financial stress cause depression, which in turn caused withdrawal? You can download this data set here.
We will first demonstrate this analysis with jamovi, the free and open statistical spreadsheet, and then in R. Feel free to skip down to the R section.
Mediation analysis with jamovi
We’ll begin by loading up the estress.csv data set into jamovi. If you haven’t installed the medmod module yet, go to the jamovi library and install it. Subsequently, select the mediation analysis from the medmod menu icon and assign the appropriate options in the options panel as follows:
You can download the jamovi file over here and open it in jamovi to get access to the data, results, and options. Before discussing the results, we’ll demonstrate how this same analysis can be done in R.
Mediation analysis with R
We’ll first read the data into our R session:
data <- read.csv("estress.csv")
head(data)
## tenure estress affect withdraw sex age ese
## 1 1.67 6.0 2.60 3.00 1 51 5.33
## 2 0.58 5.0 1.00 1.00 0 45 6.05
## 3 0.58 5.5 2.40 3.66 1 42 5.26
## 4 2.00 3.0 1.16 4.66 1 50 4.35
## 5 5.00 4.5 1.00 4.33 1 48 4.86
## 6 9.00 6.0 1.50 3.00 1 48 5.05
Next, we will perform the mediation analysis using medmod. The variables we will use for our mediation analysis are estress (economic stress), withdraw (withdrawal from entrepreneurial activities), and affect (depressed affect). If you haven’t installed the medmod package yet, install it as follows: install.packages('medmod'). I added a couple of additional options to give us some extra information.
library('medmod')
results <- med(data, dep = 'withdraw', med = 'affect',
pred = 'estress', label = TRUE, paths = TRUE,
pm = TRUE)
Interpreting results
Let’s check out the results of our mediation analysis:
##
## MEDIATION
##
## Mediation Estimates
## ─────────────────────────────────────────────────────────────────────────────────
## Effect Label Estimate SE Z p % Mediation
## ─────────────────────────────────────────────────────────────────────────────────
## Indirect a × b 0.1330 0.0288 4.62 < .001 63.4
## Direct c -0.0768 0.0521 -1.48 0.140 36.6
## Total c + a × b 0.0561 0.0540 1.04 0.299 100.0
## ─────────────────────────────────────────────────────────────────────────────────
##
##
## Path Estimates
## ──────────────────────────────────────────────────────────────────────────────
## Label Estimate SE Z p
## ──────────────────────────────────────────────────────────────────────────────
## estress → affect a 0.1729 0.0295 5.85 < .001
## affect → withdraw b 0.7691 0.1025 7.51 < .001
## estress → withdraw c -0.0768 0.0521 -1.48 0.140
## ──────────────────────────────────────────────────────────────────────────────
The first table consists of the mediation estimates while the second table consists of the individual path estimates. The Label column shows how these results are related to each other. The mediation estimates show that the indirect effect differs significantly from zero, indicating that there is indeed a mediation effect. The % Mediation column shows which percentage of the total effect is accounted for by the indirect effect and can be used as an mediation effect size. For the exact nature of the mediation effect we’ll take a look at the path estimates; economic stress increases depressed affect, and depressed affect in turn increases withdrawal from entrepreneurial activities.
Mediation analysis with lavaan
For its computations medmod uses lavaan—a powerful R package created by Yves Rosseel used to fit latent variable models. Where medmod focuses on two specific models, lavaan gives its users more freedom in their model specification. You can specify your latent variable model using lavaan model syntax. medmod tries to make it easy to transition to lavaan by providing the lavaan syntax used to fit the mediation and moderation analyses.
The lavaan model syntax is part of the medmod results object and you can simply extract it as follows:
model <- results$modelSyntax
cat(model)
## # Direct effect
## withdraw ~ c*estress
##
## # Mediator
## affect ~ a*estress
## withdraw ~ b*affect
##
## # Indirect effect (a*b)
## ab := a*b
##
## # Total effect
## total := c + (a*b)
You can now pass the model syntax into the sem function from the lavaan package, to do the exact same analysis but now directly in lavaan:
library('lavaan')
fit <- sem(data = data, model = model)
summary(fit)
## lavaan (0.5-23.1097) converged normally after 16 iterations
##
## Number of observations 262
##
## Estimator ML
## Minimum Function Test Statistic 0.000
## Degrees of freedom 0
##
## Parameter Estimates:
##
## Information Expected
## Standard Errors Standard
##
## Regressions:
## Estimate Std.Err z-value P(>|z|)
## withdraw ~
## estress (c) -0.077 0.052 -1.475 0.140
## affect ~
## estress (a) 0.173 0.030 5.853 0.000
## withdraw ~
## affect (b) 0.769 0.102 7.506 0.000
##
## Variances:
## Estimate Std.Err z-value P(>|z|)
## .withdraw 1.269 0.111 11.446 0.000
## .affect 0.461 0.040 11.446 0.000
##
## Defined Parameters:
## Estimate Std.Err z-value P(>|z|)
## ab 0.133 0.029 4.616 0.000
## total 0.056 0.054 1.039 0.299
You can also use the lavaan model syntax provided by medmod as a starting point to create more complex models (e.g., moderated mediation). For more information about lavaan, check out the official lavaan website lavaan.ugent.be.
Comments